Under the support of the National Natural Science Foundation of China (Grant Nos. 61925601 and 62006137), Professor Yang Liu's research group from the Department of Computer Science and Technology and the Institute for AI Industry Research at Tsinghua University, in collaboration with Assistant Professor Wenbing Huang from Gaoling School of Artificial Intelligence at Renmin University of China, have made progress in the field of artificial intelligence-assisted antibody design. Their research paper entitled "Conditional Antibody Design as 3D Equivariant Graph Translation" (link: http://openreview.net/pdf?id=LFHFQbjxIiP), was selected as the Outstanding Paper Honorable Mention at the Eleventh International Conference on Learning Representations (ICLR 2023), which is a leading international conference in the field of artificial intelligence.
Antibodies are Y-shaped proteins used by the immune system to identify and neutralize bacteria, viruses, and other pathogens. Antibody-based drugs have played a crucial role in the treatment of autoimmune diseases and cancers. Antibody design and optimization are extremely challenging mainly due to three aspects. First, the search space of amino acid sequences is enormous, with approximately 10 trillion possible combinations of the amino acids for each CDR, and we need to consider multiple CDRs. Second, the antibody structure is influenced by complex internal and external physical interactions, and the 3D structure formed by the interaction of amino acids in the antibody should tightly bind to the 3D structure of the antigen, especially its epitope. Finally, antibody design needs to follow the symmetry of physical laws and should not be affected by translations and rotations of the coordinate system.
The paper proposes a deep learning-based method for antibody design called Multichannel Equivariant Attentive Network (MEAN). The method smartly borrows the core idea of end-to-end neural machine translation technology, treating antibody generation as a 3D equivariant graph translation problem: given the 3D antibody-antigen complex, output the 1D amino acid sequence and docked 3D structure of CDRs. MEAN uses equivariant graph neural networks to process protein structures directly in 3D space, effectively overcoming the limitation of traditional methods that can only incorporate 3D structure information in the preprocessing stage. By fully utilizing the complete context including the information of the target antigen and the variable region of the antibody, MEAN efficiently and synchronously generates the 1D sequence and 3D structure of CDRs under the symmetry of physical laws, yielding enhanced specificity, expressivity, and generalization capabilities. In the task of antigen-binding CDR design, the amino acid reconstruction accuracy of MEAN is 23% higher than RefineGNN proposed by MIT. In the task of binding affinity optimization, the Gibbs free energy difference (ddG) of the antibodies optimized by MEAN is 34% higher than that of RefineGNN. Figure 1 (A) contrasts the difference between MEAN and RefineGNN, where the CDR amino acid sequence generated by MEAN is "ANGDGDY", which largely overlaps with the true sequence "ANWAGDY", and the corresponding predicted 3D structure is almost identical to the ground-truth structure. Figure 1 (B) shows the affinity distribution of the optimized antibodies for the target antigen using both methods. Compared to the original antibody (RAbD), the affinity of the antibodies optimized by MEAN is remarkably improved (smaller value indicates better result), surpassing RefineGNN significantly.
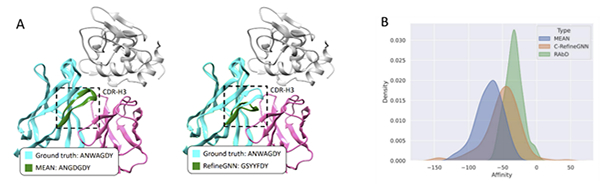
Figure 1. Comparisons between MEAN and RefineGNN. (A) CDR-H3 comparison. The gray part in the figure represents the antigen, and the green and red parts represent the heavy and light chains of the antibody. The bottom left corner shows the ground-truth amino acid sequence and the generated amino acid sequence. The dashed box in the figure denotes the generated 3D structure of CDR-H3. (B) Affinity comparison. The green area in the figure is the density distribution curve of the original antibody's affinity for the target antigen in the dataset, and the orange and blue areas represent the density distribution curves of the antibody's affinity after optimization by RefineGNN and MEAN, respectively.

Add: 83 Shuangqing Rd., Haidian District, Beijing, China
Postcode: 100085
Tel: 86-10-62327001
Fax: 86-10-62327004
E-mail: bic@donnasnhdiary.org
京ICP备05002826号 文保网安备1101080035号 Copyright 2017 NSFC, All Right Reserved